|
Once the feature and content of an image have been charaterized,
KMeD clusters the extracted image content based on the
different features (i.e. the attributes of the tumors). The clustering technique
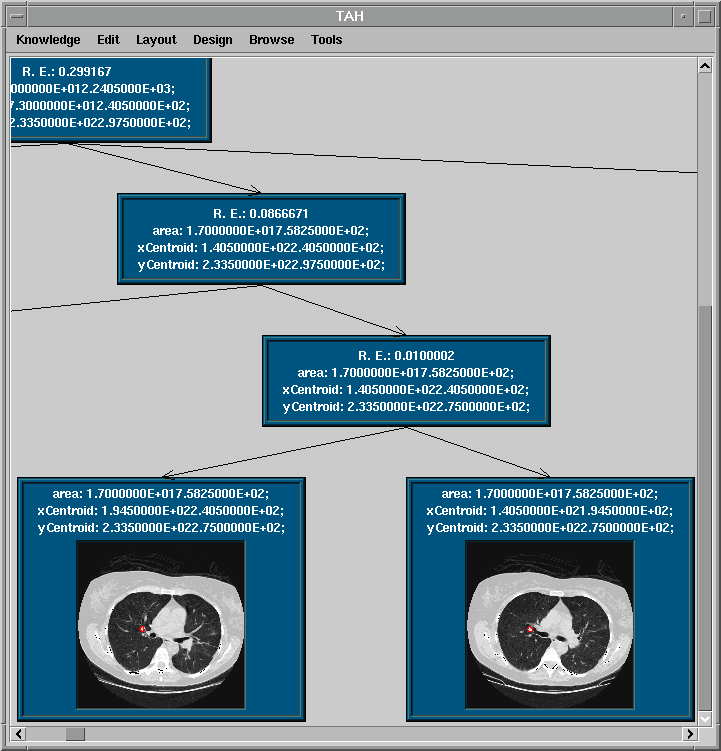
is based on the Type Abstraction Hierarchy (TAH) model where similar
objects are clustered together based on the distribution of data. By using the TAH, we are able to take
advantage of the generalization/specialization properties inherent in the TAH structure. The generalization (specialization) property of the TAH allows us to relax (constrain) the predicates of a query in the event that more (less) results for query are required.
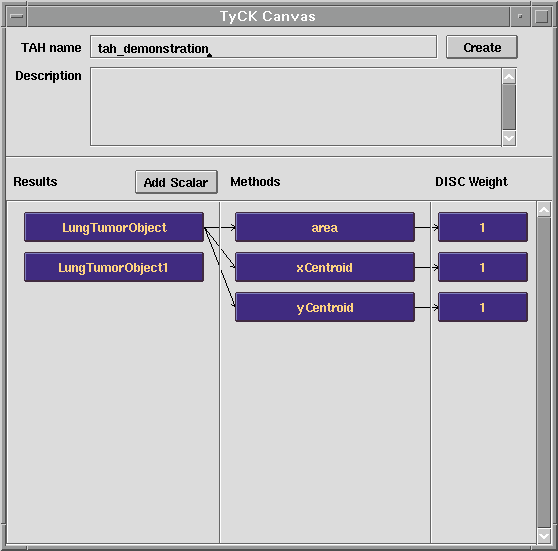
To construct a TAH, KMeD uses the Type Abstraction Hierarchy Construction Kit (TyCK) tool, shown in the next figure. The items in the left most column correspond to the 'content' (e.g. sub-image objects) that are stored in the database; in the figure displayed below, these objects refer to lung tumors. The second column specifies the user-specified features (or attributes) of these lung tumor objects that the TyCK module will use to cluster the lung tumor objects in our database during TAH construction. The last column of TyCK allows the user to associate weights with the chosen attributes.
|