|
|
|
|
|
|
|
|
 |
||||||
| A part of the National Science Foundation Scientific
Database Initiative Grant IRI 9116849 Last updated on August 17th, 1998 |
|
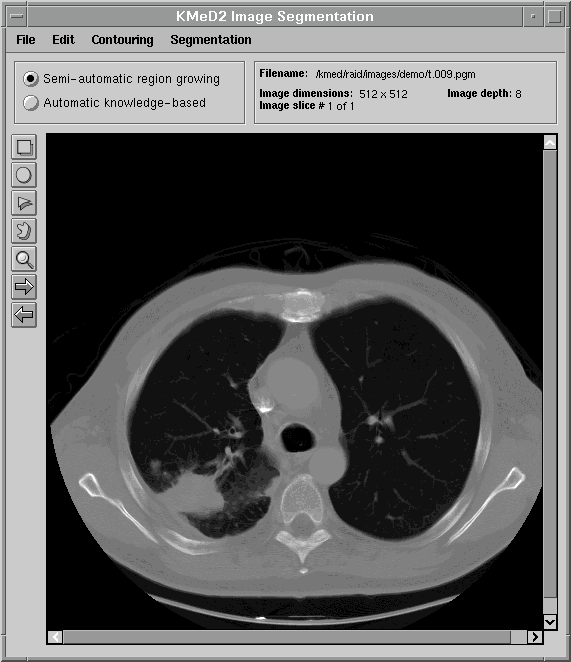
One of the goals of KMeD is to support the retrieval of images by content and feature. Because the domain currently supported by KMeD is that of radiology, the concepts of 'content' and 'feature' will be defined within the context of this area of medicine. However, the functionality of KMeD is not confined by nor limited to the medical arena. In order to support the querying of images by feature and content, the 'content' must first be extracted from the images. An example of this 'content' is a tumor that is visible in a lung image (as is shown below). KMeD provides tools that allow its users to segment sub-images (or 'content') from their parent image. The extracted result forms the contour of the sub-image. From this contour, features of the sub-image, such as area and perimeter, can then be calculated. This extracted information is stored in the KMeD database and may then be queried by KMeD users.
The next three figures demonstrate how KMeD's segmentation tool uses a region of interest (ROI), supplied the user, to determine the contour of an object.
The first figure displays a typical lung image, a slice taken from a series of lung CT scans.
 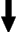 The user then draws a region of interest using various annotation tools. In this figure, the user has chosen to use the freehand drawing tool to outline a tumor in the lower-left of the lung image. 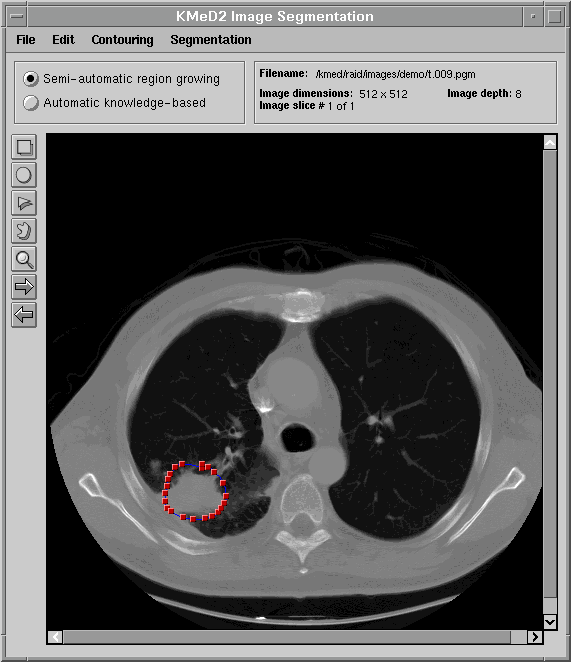 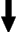 Using the outline provided above, KMeD's segmentation module computes a more exact contour of the tumor region. This contour is then saved in our database in the form of a list of coordinate points. From this set of contour points, various features of the lung tumor object, such as area and perimeter, can then be calculated. 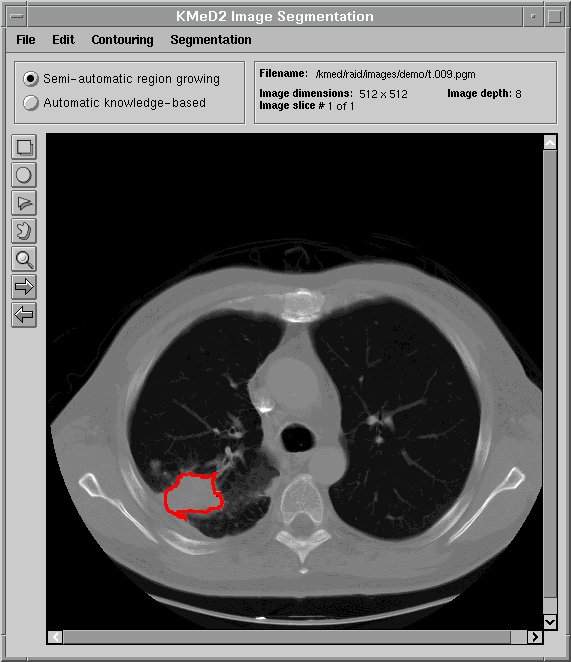 |
|
|
|
| Return to the previous KMeD page |