|
|
|
|
|
|
|
|
 |
||||||
|
The KMeD project has completed. The KMeD website now serves as a historical record of the findings and accomplishments of the KMeD team. A part of the National Science Foundation Scientific Database Initiative Grant IRI 9116849 Last updated on June 12th, 2001 |
|
KMeD's query language is KMeD PICQUERY+, a menu-driven, tabular language. Key features include:
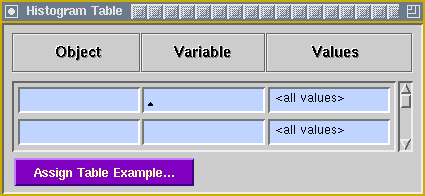
Common domain-specific queries can be encapsulated into a custom, user-specified application. The application serves as a shell that accepts the user's specifications, then builds a KMeD PICQUERY+ query based on those instructions. The application also provides access to relevant parts of the underlying data model and knowledge base. Predicate TablesThe screenshot below illustrates a sample KMeD PICQUERY+ predicate table; click on it to see the menu that pops up for the Relation Operator column of the table. Processing TablesThe screenshot below illustrates a sample KMeD PICQUERY+ processing table. In particular, this table is used for building histograms from retrieved query results.
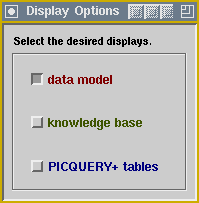
Custom ApplicationsThe screenshot below displays KMeD's prototype patient finder application, including a menu that hides or shows windows for the application's data model, knowledge base, and KMeD PICQUERY+ tables. Click on the menu to view a sample knowledge base display. 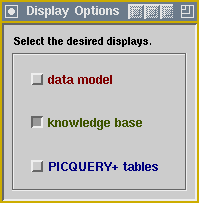
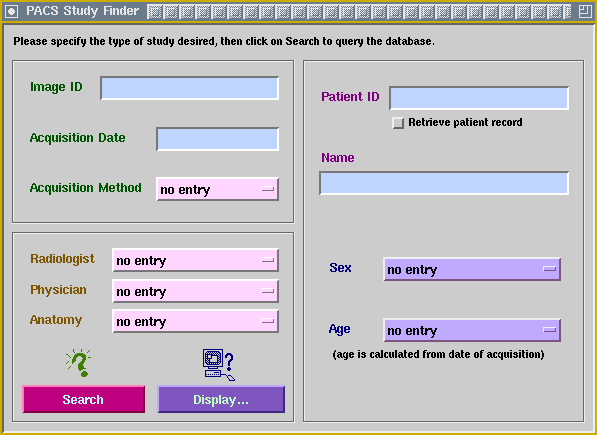
Below is a screenshot of another custom application, KMeD's prototype PACS study finder. Its accompanying display menu has the application's data model selected; click on it to see what the data model looks like.
|
|
|
| Return to the previous KMeD page |